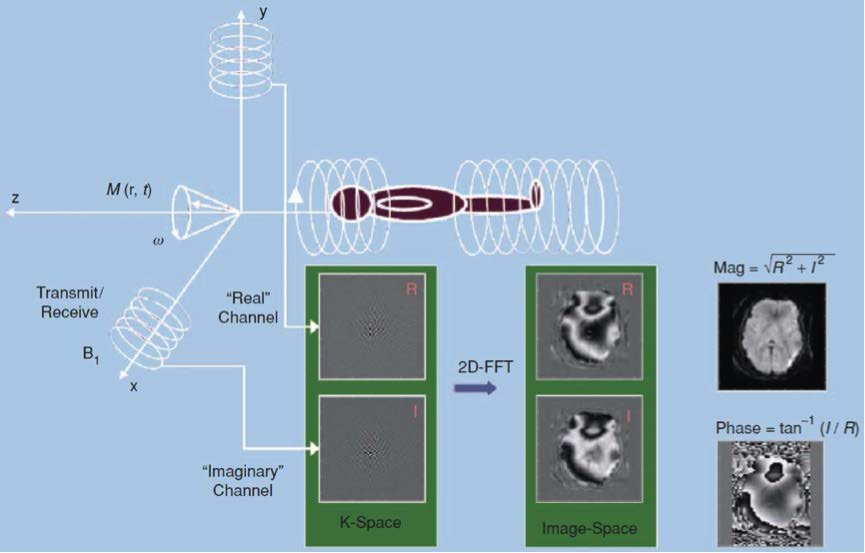
 Source: V.d. Calhoun and T. Adali (2012)
Source: V.d. Calhoun and T. Adali (2012)
Bayesian Spatial Modeling via Kernel Convolutions on Complex-Valued fMRI Signals
Abstract
We propose Bayesian variable selection tools coupled with a novel spatial kernel convolution for detecting brain activation at the voxel level in complex-valued fMRI (CV-fMRI) data. We develop a computationally efficient MCMC algorithm within this modeling framework that takes advantage of kernel-based dimension reduction. The proposed spatial model leads to a reduction of false positives when compared to complex-valued models that do not incorporate spatial structure. The kernel-based model leads to more accurate posterior probability activation maps, and hence less false positives than alternative spatial approaches based on Gaussian process models. We also find that the complex-valued approaches dominate magnitude-only approaches when the signal-to-noise-ratio (SNR) and the contrast-to-noise ratio (CNR) are low. Furthermore, we compare spatial and non-spatial magnitude-only models and find that the kernel-based spatial approach considerably improves the sensitivity when detecting activation at the voxel level. We analyze simulated and human task-related CV-fMRI data and show that the proposed Bayesian complex-valued kernel-based model leads to more precise activation maps and more accurate activation results at the voxel level than those obtained from complex-valued models with alternative spatial and non-spatial structures.
More detail can easily be written here using Markdown and $\rm \LaTeX$ math code.